News
Molecular Microbiology
Bacterial motility depends on a critical flagellum length and energy-optimized assembly
-
In our latest study, we reveal how bacteria optimize the assembly of their flagella to achieve motility as efficiently as possible. Using synchronized flagellar growth in Salmonella and a combination of live-cell imaging, electron microscopy, and biophysical modeling, we determined that a critical filament length of ~2.5 μm is necessary to trigger swimming. This threshold reflects an elasto-hydrodynamic instability that enables propulsion once the filament is long enough to overcome the stabilizing elasticity of the flagellar hook. Remarkably, we measured flagellin secretion rates of up to 10,000 amino acids per second, placing the flagellar type-III secretion system (fT3SS) among the fastest protein exporters known. Our model shows that this high rate is not just a byproduct of the system but is evolutionarily optimized to minimize the energy cost per unit of motility gained. Beyond this threshold, increasing secretion speed yields diminishing returns in swimming performance but significantly increases energy expenditure. These findings highlight a precise evolutionary tuning of secretion dynamics and mechanical properties to ensure rapid and efficient motility. Published in PNAS!
Structure and mechanism of the Zorya anti-phage defence system
-
In our latest study, now published in Nature, we reveal the structure and mechanism of the widespread anti-phage Zorya defense system in bacteria. We show that ZorA and ZorB form a membrane-anchored, proton-driven rotary motor that senses phage-induced envelope perturbations and transmits a mechanical signal via a long intracellular tail. This signal activates the soluble effectors ZorC and ZorD, which bind and degrade phage DNA directly—without triggering abortive infection or host cell death. Using cryo-EM, functional assays, and mutational analyses, we demonstrate that all Zorya components are essential for defense, and that ZorAB resembles the bacterial flagellar stator both structurally and mechanistically. The motor’s activation depends on peptidoglycan anchoring and proton flow, while the tail coordinates effector recruitment. ZorC exhibits DNA-binding activity, and ZorD acts as a latent nuclease, rapidly degrading phage genomes upon activation. This work defines Zorya as a mechanically triggered, membrane-proximal antiviral system and illustrates how bacteria can leverage motor proteins for direct, localized immunity.
FlhE functions as a chaperone to prevent formation of periplasmic flagella in Gram-negative bacteria
-
In our new study published in Nature Communications, we uncover a critical role for the periplasmic protein FlhE in ensuring proper flagellar assembly in Salmonella enterica. We show that FlhE prevents the aberrant formation of periplasmic flagella by acting as a chaperone during the rod assembly stage of the flagellar basal body. In the absence of FlhE, premature assembly of hook and filament subunits occurs within the periplasm, disrupting peptidoglycan synthesis and cell division machinery. This leads to abnormal cell morphology, inner and outer membrane stress, and ultimately cell lysis. Combining live-cell super-resolution microscopy, genetic perturbations, and electron microscopy, we demonstrate that FlhE stabilizes the interaction between the rod-cap FlgJ and the distal rod until the outer membrane-associated L-ring is assembled. Our findings suggest that FlhE fine-tunes the timing of the flagellar substrate specificity switch to safeguard the spatial integrity of flagellum morphogenesis. This work not only resolves a long-standing question about FlhE function but also highlights a novel mechanism by which bacteria prevent lethal misassembly of complex nanomachines.
DNA transfer during bacterial conjugation uses the F pilus as conduit
-
Horizontal transfer of mobile genetic elements by bacterial conjugation is responsible for the spread of antibiotic resistance and virulence among pathogenic bacterial species, a growing global health problem. In a collaborative effort with the team of Christian Lesterlin, we investigated the role of the F pilus in bacterial conjugation. Utilizing advanced live-cell microscopy and innovative fluorescent reporter systems, we demonstrate that the F pilus can act as a conduit for DNA transfer between physically distant bacteria, in addition to transfer between cells in direct contact. This finding revises our understanding of the mechanism of bacterial conjugation and its impact on the dissemination of drug resistance and virulence genes in bacterial communities. Published in PNAS!
See below for a time lapse movie of the F pilus labelled with AF-Mal488 showing one extension (top) and one retraction (bottom) event. Scale bar 1 μm and time in minutes are indicated (10 sec/frame). The second time-lapse movie shows distant transfer events between E. coli cells. Distant transfer is reported by both the formation Ssb-Ypet conjugative foci at each edge of the pilus connecting the mating pair cells and the subsequent the formation of mCherry-ParB foci, which confirms the acquisition of the dsDNA plasmid by the recipient cell. Three different events are shown. Scale bar 1 μm and time in minutes are indicated (3 min/frame).
Gita Naseri awarded a six-year grant by the Emmy Noether Program of the German Research Foundation (DFG)
 Dr. Gita Naseri (Emmanuelle Charpentier’s laboratory at the Max Planck Unit for the Science of Pathogens (MPUSP) and Marc Erhardt’s laboratory at the Institute for Biology, Humboldt-Universität zu Berlin) has been awarded a six-year grant by the Emmy Noether Program of the German Research Foundation (Deutsche Forschungsgemeinschaft (DFG))
Dr. Gita Naseri (Emmanuelle Charpentier’s laboratory at the Max Planck Unit for the Science of Pathogens (MPUSP) and Marc Erhardt’s laboratory at the Institute for Biology, Humboldt-Universität zu Berlin) has been awarded a six-year grant by the Emmy Noether Program of the German Research Foundation (Deutsche Forschungsgemeinschaft (DFG))
The Emmy Noether Programme has been established by the DFG to offer exceptionally qualified early career researchers the chance to qualify for a professorship at a university by leading an independent junior research group for a period of six years. The Emmy Noether grant awarded to Gita Naseri entitled „COMPLATn - A comprehensive platform for sustainable biosynthesis of rare natural products in Pichia pastoris” aims to create a new avenue for the biomanufacturing of rare natural products that control microbial diseases and cancers in microbial cells by employing cutting-edge genetic engineering technologies. For this project, Gita Naseri is receiving a total funding of almost 2.6 million EUR over a 6-year period.
After studying biology in Iran, Gita Naseri received her doctoral degree in synthetic biology from the University of Potsdam in 2018. In 2019, she embarked on her PostDoc research journey at the Institute for Chemistry at Humboldt-Universität zu Berlin. In 2021, she joined Marc Erhardt's laboratory at the Institute for Biology, Humboldt-Universität zu Berlin. Since then, she has been working as a PostDoc at Emmanuelle Charpentier’s laboratory at MPUSP and the Erhardt laboratory at the Institute for Biology of the Humboldt-Universität zu Berlin. Gita Naseri’s work has been recognized by several awards, including the NNPDF Fellowship award from the National Niemann-Pick Disease Foundation (USA) and the Go-Bio initial funding program of the German Ministry for Education and Research (Bundesministerium für Bildung und Forschung (BMBF), Germany).
Ion selectivity and rotor coupling of the Vibrio flagellar sodium-driven stator unit
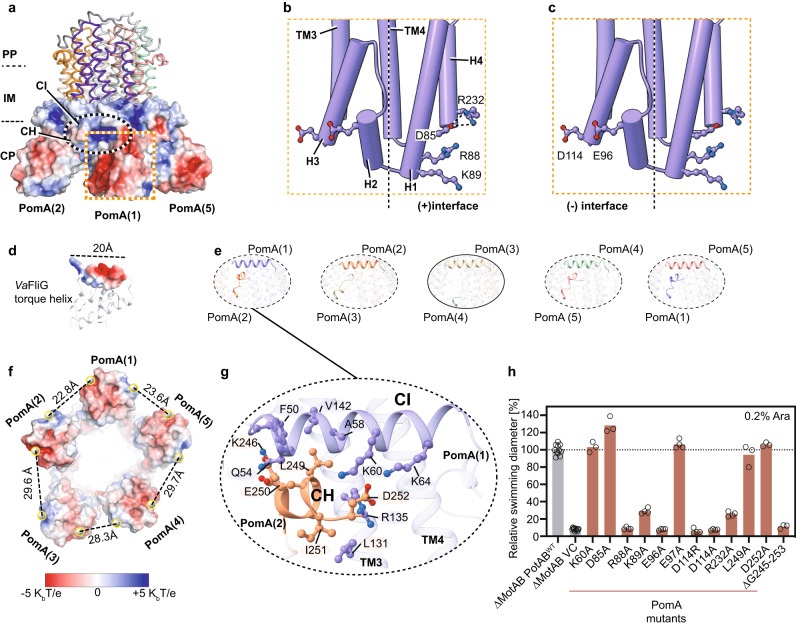 In a great collaboration with the group of Nicholas Taylor, we present a new study that revealed the detailed structure of the stator unit in the flagella of Vibrio bacteria, responsible for several human diseases. This breakthrough, achieved through cryo-electron microscopy and mutational studies, offers insights into how these bacteria use sodium ions to power movement, a key step in understanding their pathogenic behavior. Published in Nature Communications!
In a great collaboration with the group of Nicholas Taylor, we present a new study that revealed the detailed structure of the stator unit in the flagella of Vibrio bacteria, responsible for several human diseases. This breakthrough, achieved through cryo-electron microscopy and mutational studies, offers insights into how these bacteria use sodium ions to power movement, a key step in understanding their pathogenic behavior. Published in Nature Communications!
In our study, we have made significant advances in understanding the stator unit of Vibrio bacteria's flagellar motor through a combination of structural analysis, mutational studies, and molecular dynamics simulations. We have meticulously validated our cryo-electron microscopy structural results with extensive mutagenic analysis, providing robust evidence for our findings. This approach has also allowed us to propose a model for the asymmetric arrangement of the stator unit's cytoplasmic domain, which plays a crucial role in torque generation and the assembly and disassembly mechanism of the stator unit into the rotor.
See below our propsed model for the mechanisms of ion selectivity and rotor incorporation in the bacterial flagellum.
PLOS Pathogens paper: SPI-1 virulence gene expression modulates motility of Salmonella June 14, 2023
SPI-1 virulence gene expression modulates motility of Salmonella Typhimurium in a proton motive force- and adhesins-dependent manner
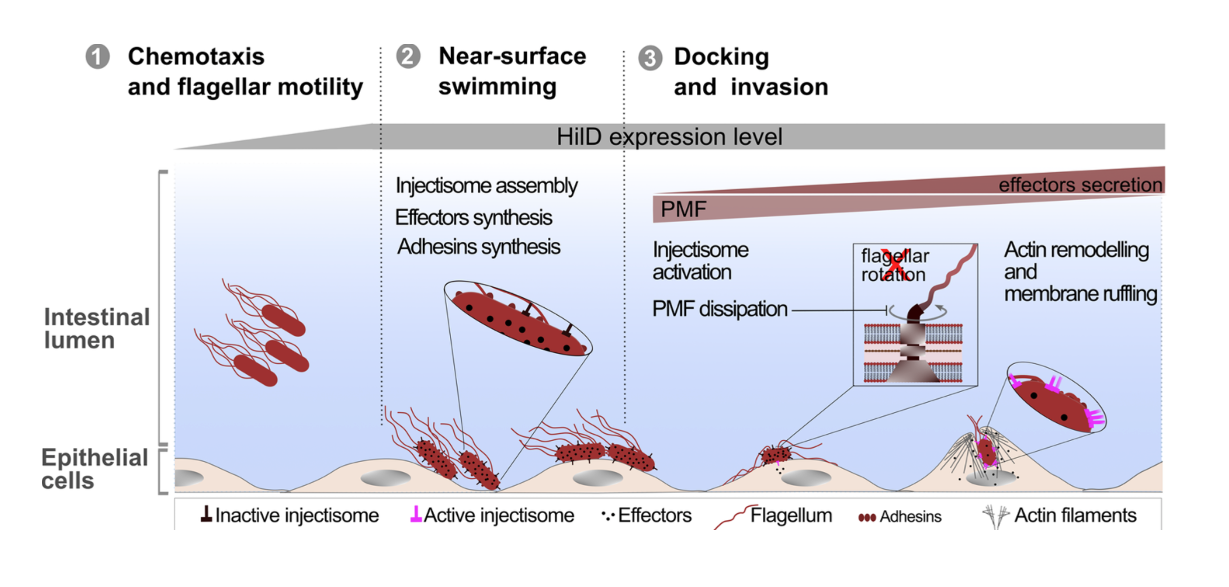
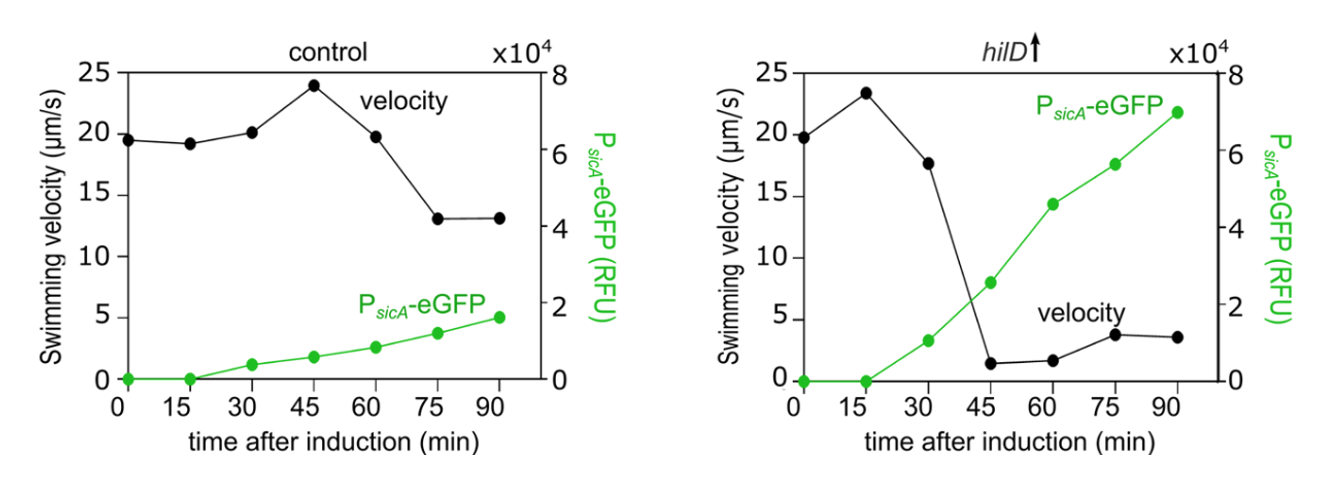 In collaboration with the groups of Enrico Klotzsch and Till Strowig, we uncover the physiological significance of the cross-talk between two main virulence factors of Salmonella, the bacterial flagellum and the Salmonella pathogenicity island-1 (SPI-1) encoded injectisome. Published in PLOS Pathogens!
In collaboration with the groups of Enrico Klotzsch and Till Strowig, we uncover the physiological significance of the cross-talk between two main virulence factors of Salmonella, the bacterial flagellum and the Salmonella pathogenicity island-1 (SPI-1) encoded injectisome. Published in PLOS Pathogens!
We found that induction of HilD, the master regulator of the SPI-1 injectisome unexpectedly leads to a significant reduction in Salmonella's motility, despite its role in activating the transcription of the flagellar master regulator: FlhDC. This drastic loss of motility is linked to the presence of SPI-1 and is independent of HilD transcriptional activation of flhDC.
See below our propsed model.
Structure and Function of Stator Units of the Bacterial Flagellar Motor
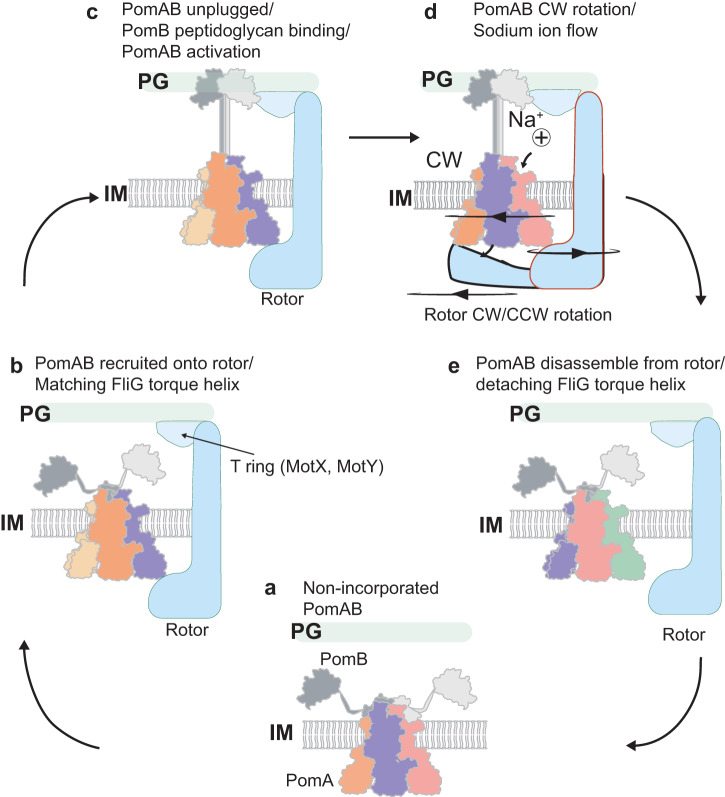
 In a great collaboration with the group of Nicholas Taylor, we present the structure of nature’s smallest rotary motor - the stator units that power bidirectional rotation of the bacterial flagellum. Published in Cell!
In a great collaboration with the group of Nicholas Taylor, we present the structure of nature’s smallest rotary motor - the stator units that power bidirectional rotation of the bacterial flagellum. Published in Cell!
The stator units (formed by the MotA–MotB membrane protein complex) use energy derived from the proton gradient across the inner membrane to power rotation of the flagellum. However, despite decades of research, their structure and molecular mode of action has remained a mystery.
In this paper, we found that the stator unit consists of a dimer of MotB surrounded by a pentamer of MotA, which interacts with the rotor of the flagellum. Quite surprisingly, the stator units function as miniature rotary motors themselves, where a pentamer of MotA rotates around a MotB dimer.
See below our propsed model.
ERC consolidator grant for BacNanoMachine
 Very happy and honored to receive funding for our project BacNanoMachine from the European Research Council!
Very happy and honored to receive funding for our project BacNanoMachine from the European Research Council!
In the project BacNanoMachine, we aim to obtain a holistic understanding of the underlying principles that allow bacteria to control and coordinate the simultaneous self- assembly processes of several multi-component nanomachines within a single cell.
Despite being unicellular organisms of relatively small size, bacteria produce sophisticated nanomachines with a high degree of self-organization. The motility organelle of bacteria, the flagellum, is a prime example of complex bacterial nanomachines. Flagella are by far the most prominent extracellular structures known in bacteria and made through self-assembly of several dozen different kinds of proteins and thus represents an ideal model system to study sub-cellular compartmentalization and self-organization.
The flagellum can function as a macromolecular motility machine only if its many building blocks assemble in a coordinated manner. However, previous studies have focused on phenotypic and genetic analyses, or the characterization of isolated sub-components. Crucially, how bacteria orchestrate the many different cellular processes in time and space in order to construct a functional motility organelle remains enigmatic.
In the project BacNanoMachine, we will combine for the first time the visualization of the dynamic self-assembly of individual flagella with quantitative single-cell gene expression analyses, re-engineering of the genetic network and biophysical modeling in order to develop a biophysical model of flagella self-assembly.
Press release


